Harnessing Graph Representations in Structural Bioinformatics
Raum 5 (E010)
Studiengang / Lehrstuhl / Firma
Bioinformatik Hochschule Mittweida and BIOTEC TU Dresden
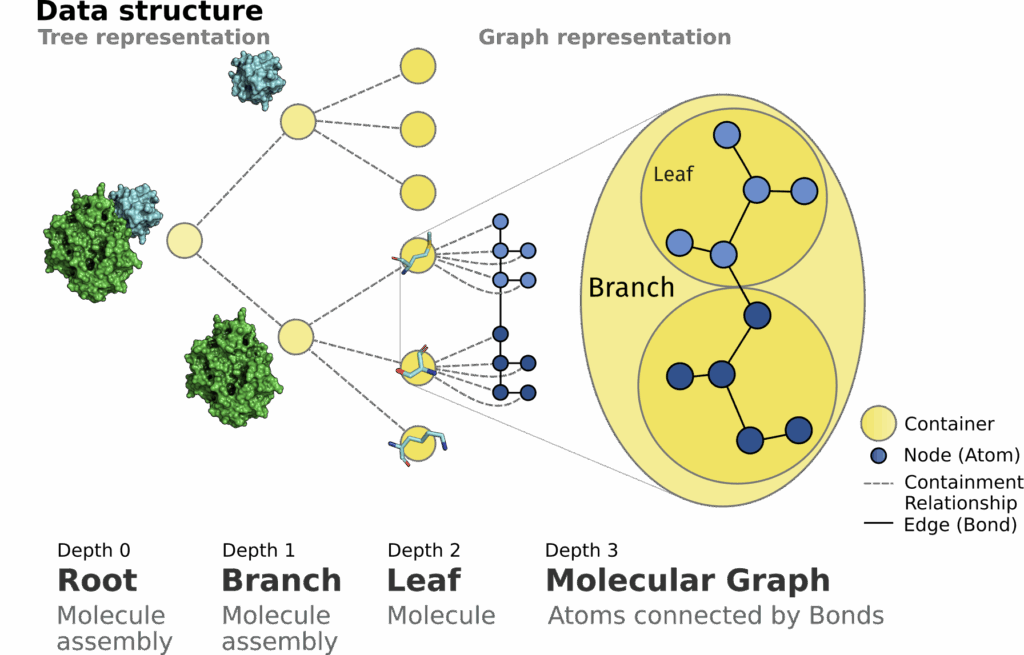
Molecules are three-dimensional structures that consist of atoms connected by covalent bonds. The application and development of algorithms to analyze their structure on a geometrical level as well as their biological implications is the focal point of structural bioinformatics. Commonly libraries store those structures as list representations of atoms. From a different point of view, the nature of molecules suggests the their modeling as graph structures. This could allow for a more efficient traversal from macro to atomic scale. We introduce the SiNGA Framework that uses graph motivated data structures to store and represent an assembly of molecules. SiNGA implements commonly used features to retrieve, investigate and work with macromolecular structures, that can be accessed using a convenient method chaining API for most features.